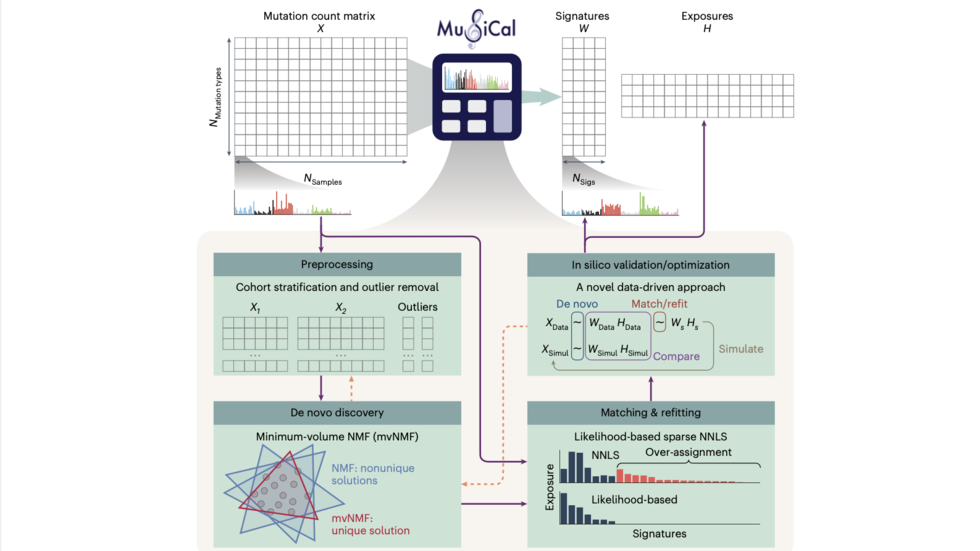
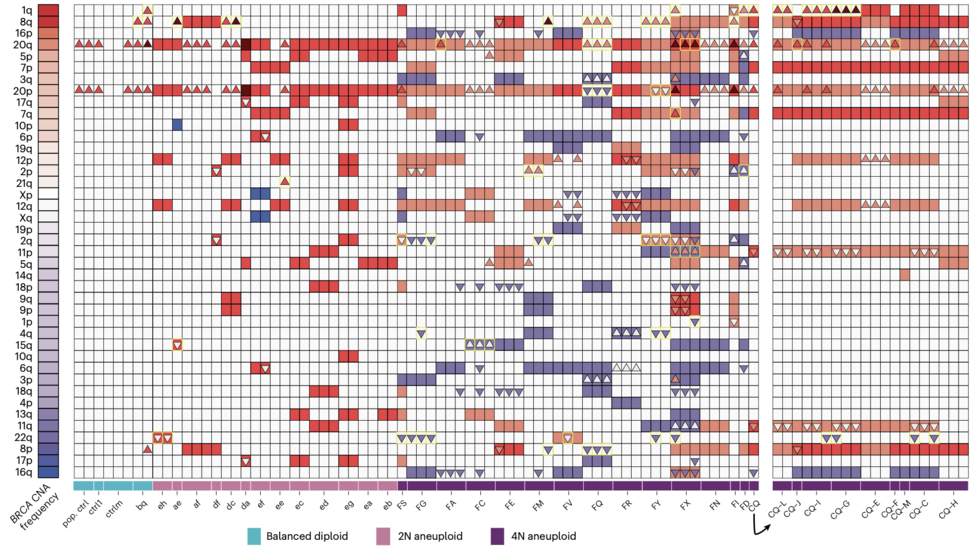
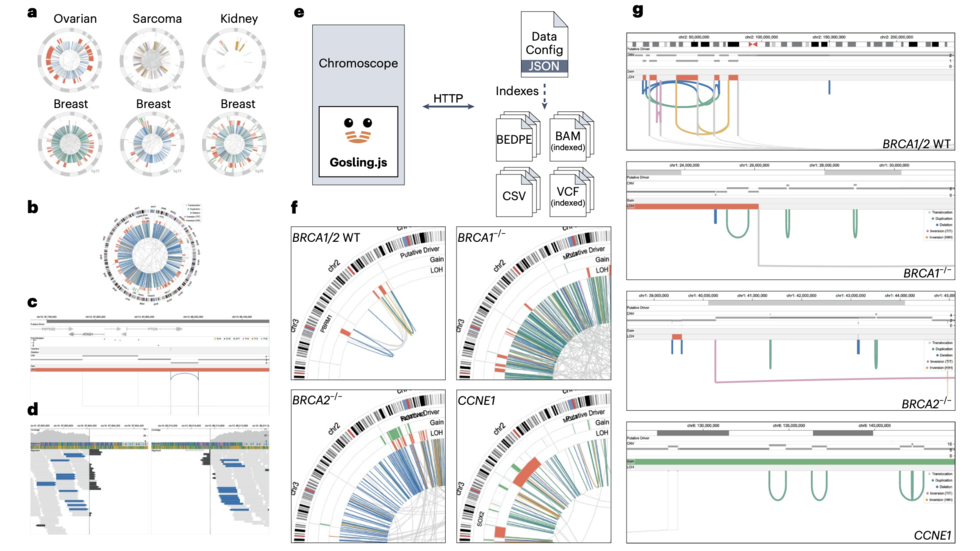
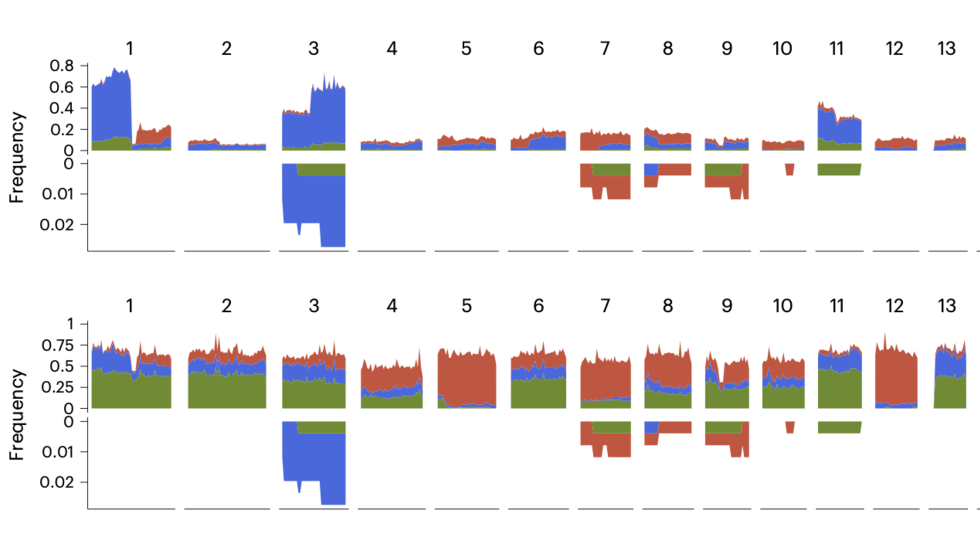
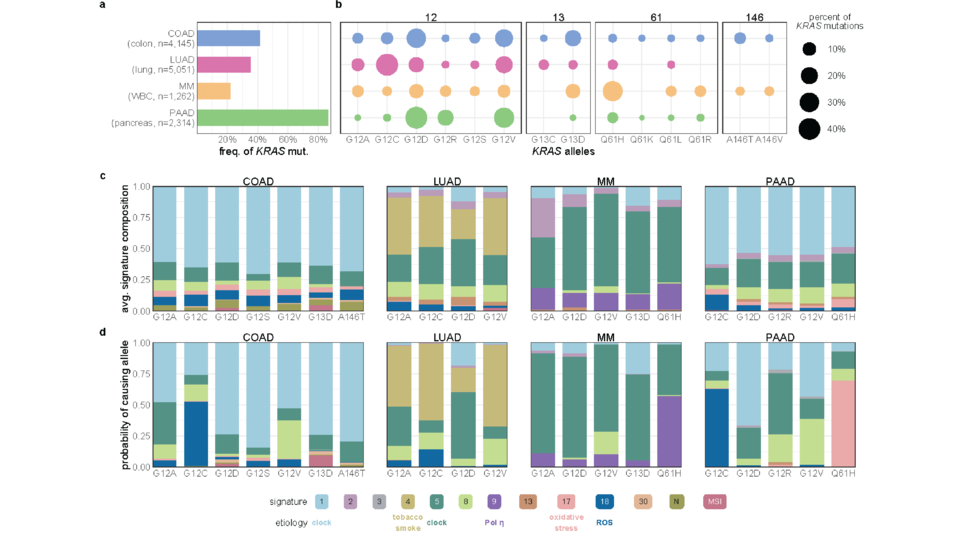
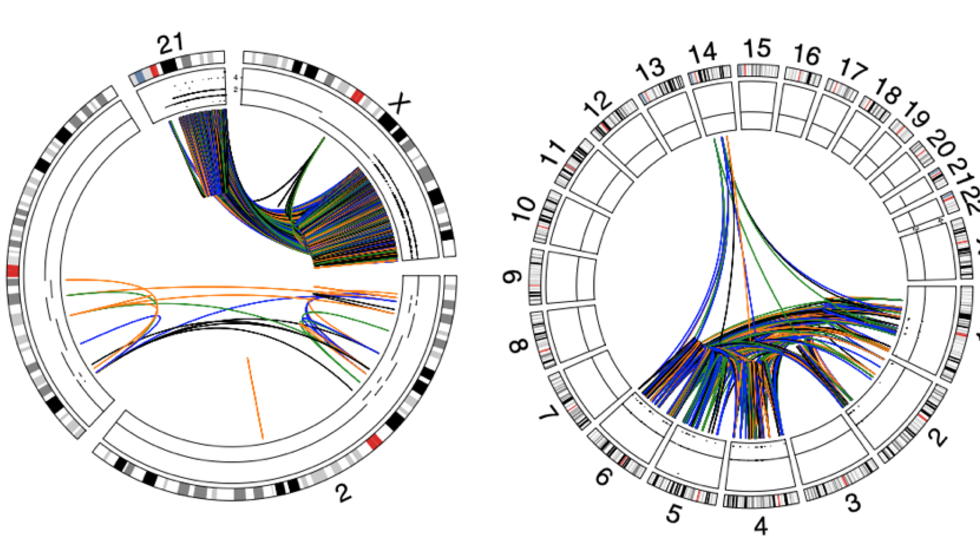
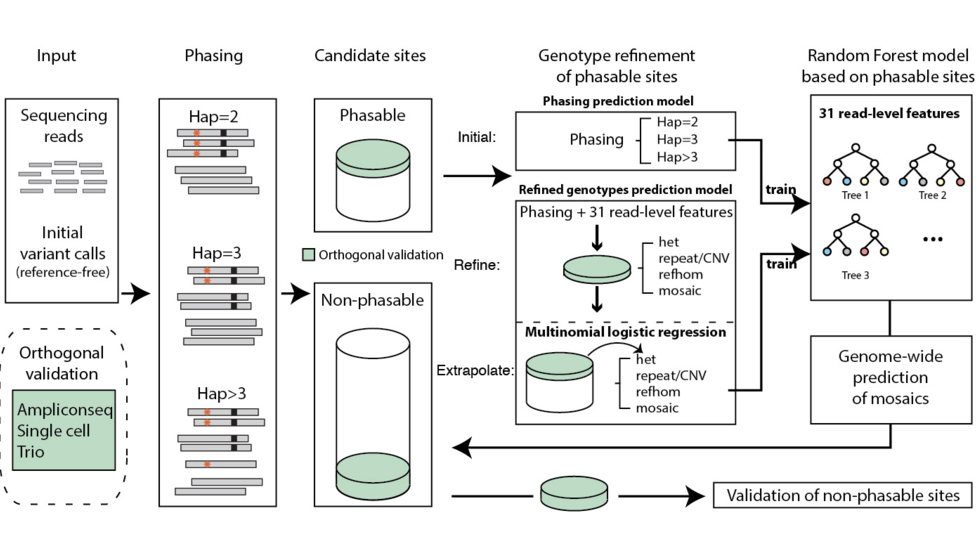
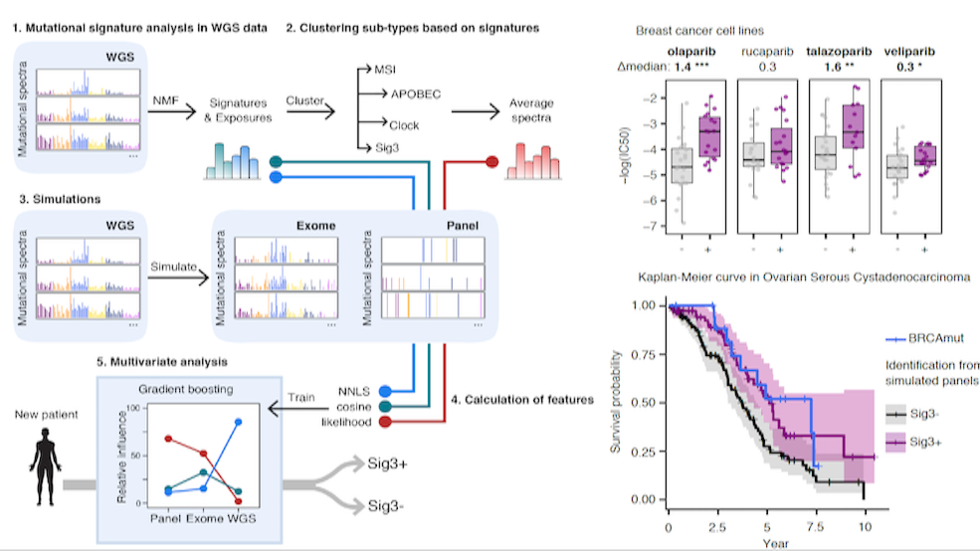
We are a bioinformatics research group in the Department of Biomedical Informatics at Harvard Medical School. We are part of the Ludwig Center at Harvard, Division of Genetics at Brigham and Women's Hospital and Harvard-MIT Division of Health, Science, & Technology.
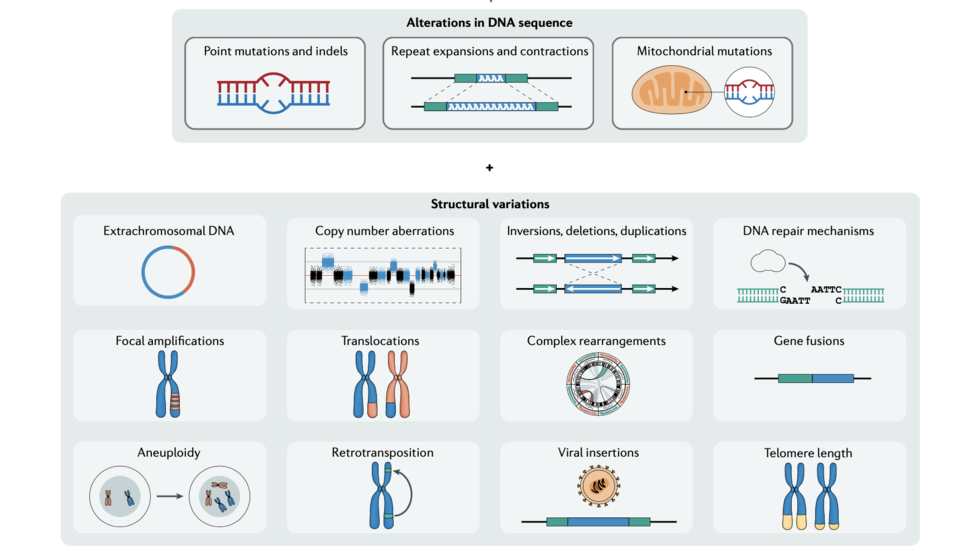
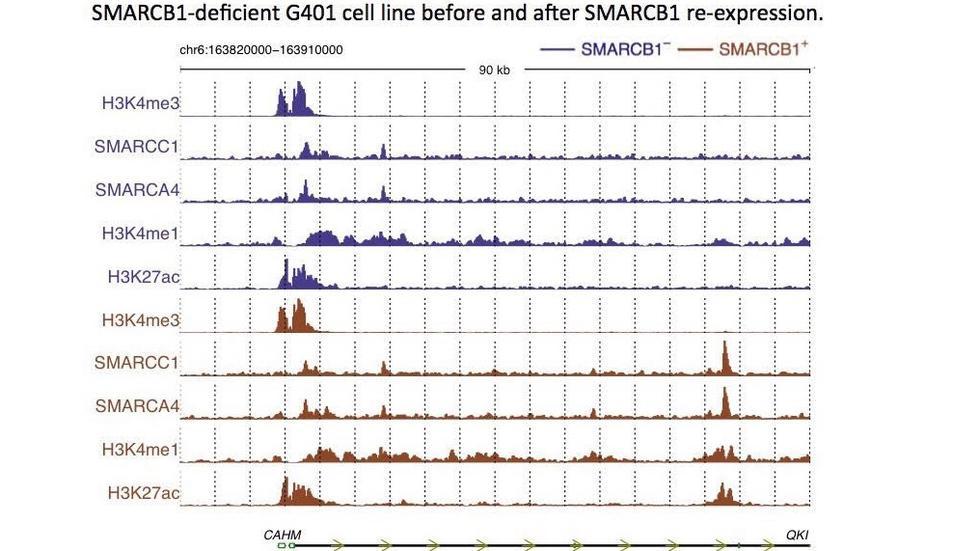
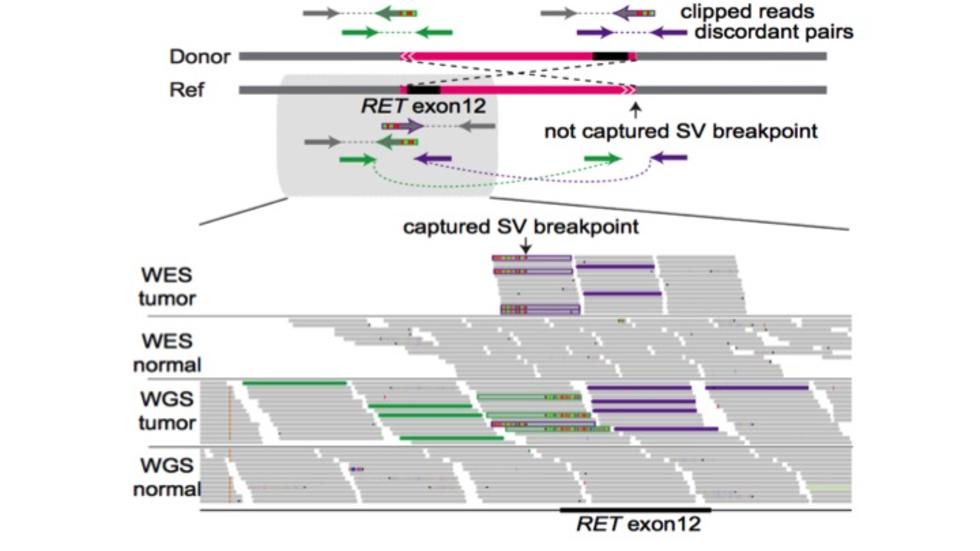
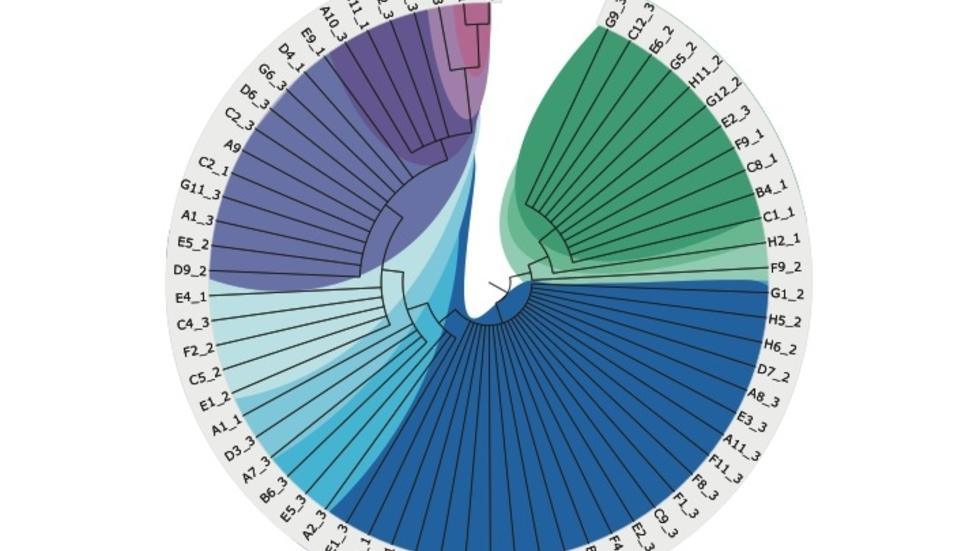
Our main aim is to understand the genetic and epigenetic mechanisms that underlie disease processes through computational and statistical analysis of genomic data. We are particularly interested in mutational processes in normal and cancer cells and their impact in gene regulation.
Collaborative scienceWe have the privilege of collaborating with a number of superb experimental laboratories at Harvard Medical School and its affiliated hospitals, as well as around the world. Our recent and current collaborators include Connie Cepko, George Daley, Steve Elledge, Konrad Hochedlinger, Mark Johnson, Mitzi Kuroda, Jordan Kreidberg, Bob Kingston, William Pu, Charlie Roberts, Chris Walsh, Fred Winston.
Here are some consortium projects in which we are involved:
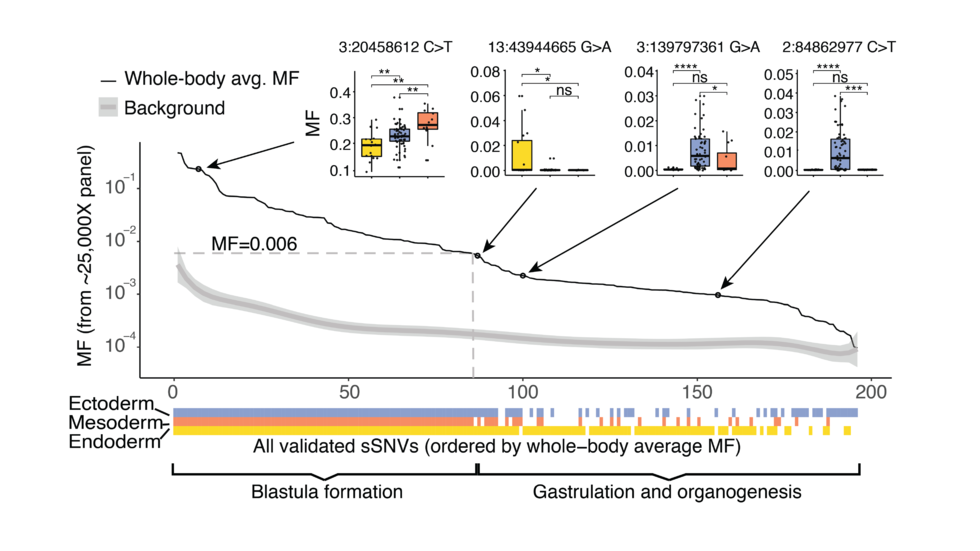
- SMaHT (Somatic Mosaicism across Human Tissues), starting in 2023 - Do mutations accumulate in non-cancer cells? Where and how? Can those mutations lead to cancer or other diseases?
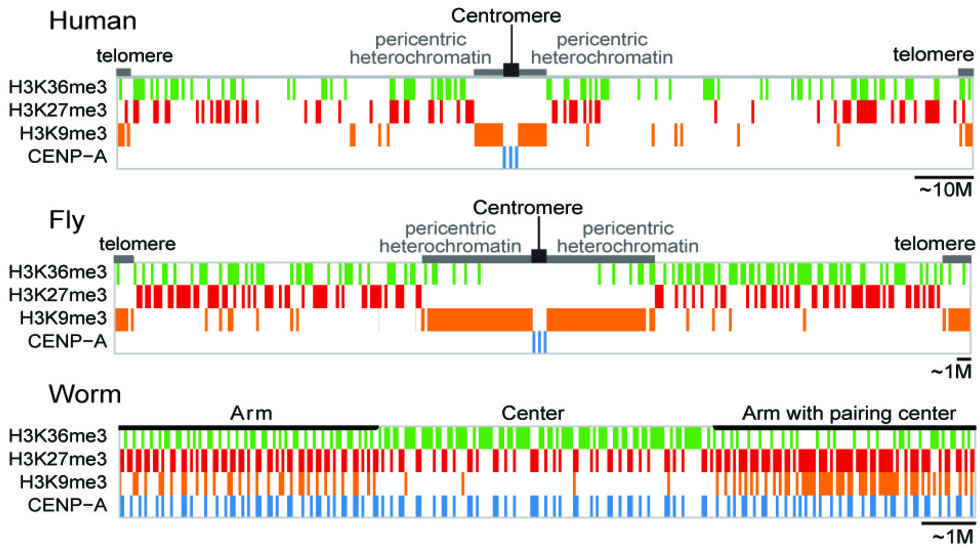
- 4D Nucleome - What are the principles underlying three-dimensional organization of the human genome? What is the role of the nuclear architecture in gene regulation? As the Data Coordination and Analysis Center for this major NIH initiative, we will collect, process, and display all data generated in the consortium and to perform integrative analysis. Main collaborators: Peter Kharchenko (HMS), Leonid Mirny (MIT), Nils Gehlenborg (HMS), Hanspeter Pfister (Harvard) and Ting Wang (WashU).
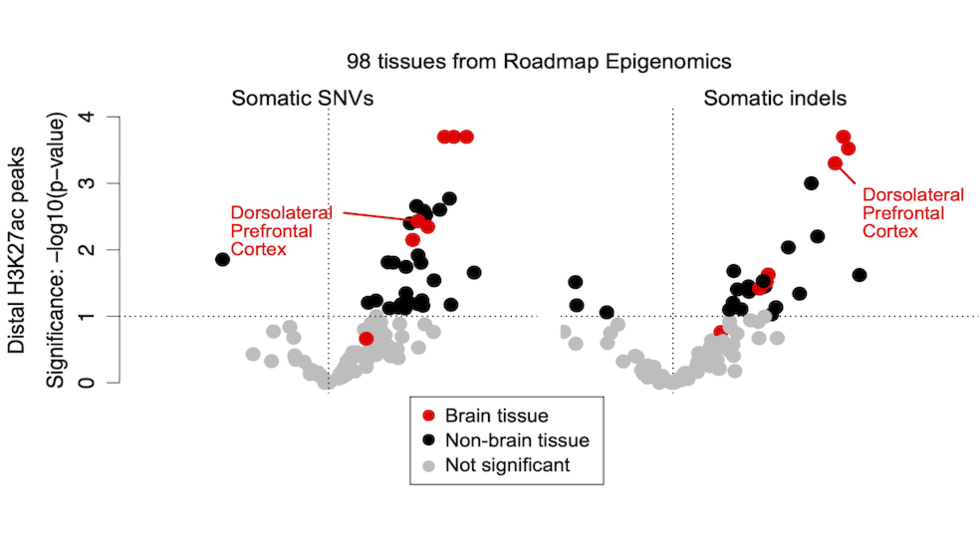
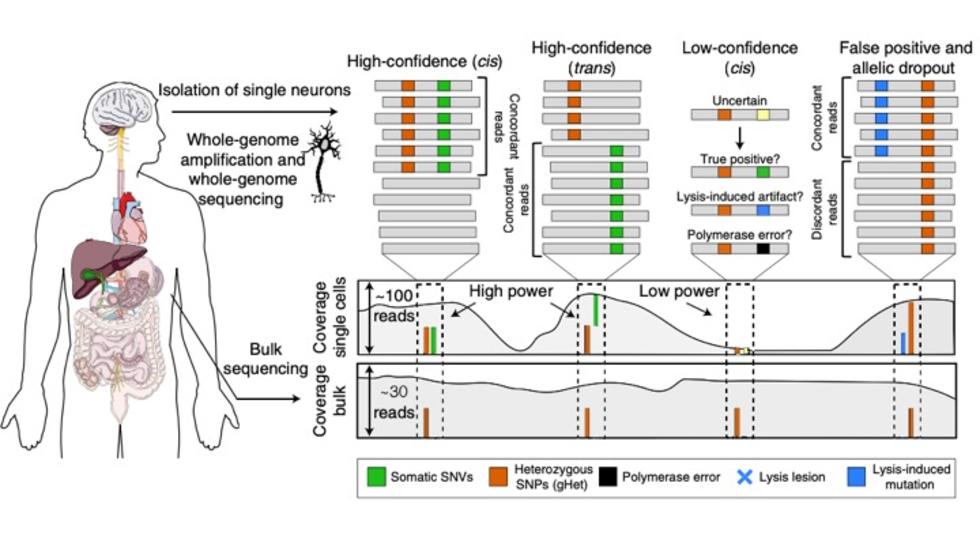
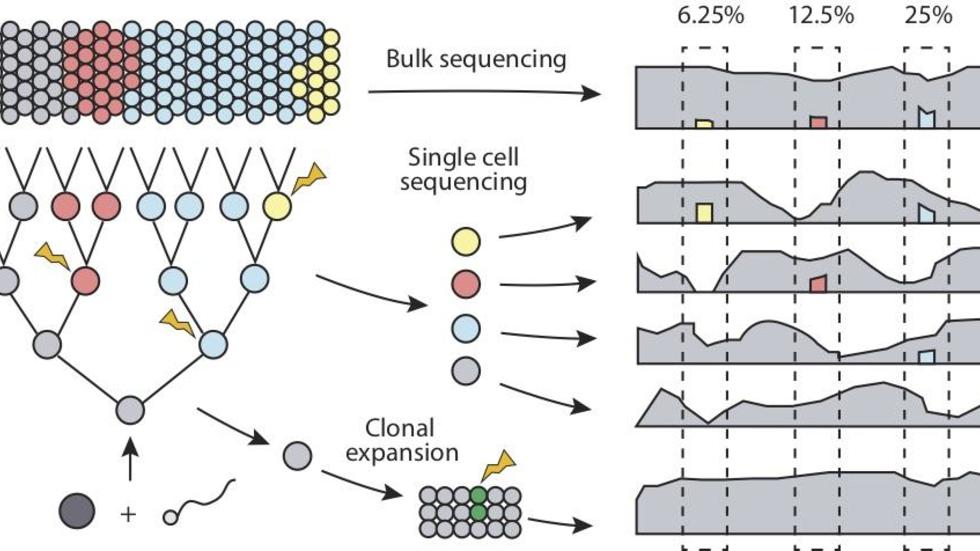
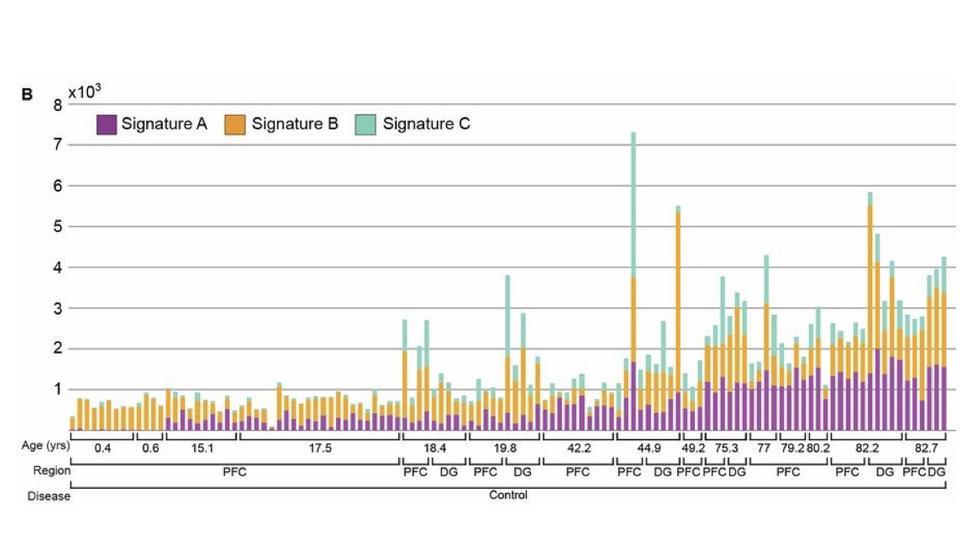
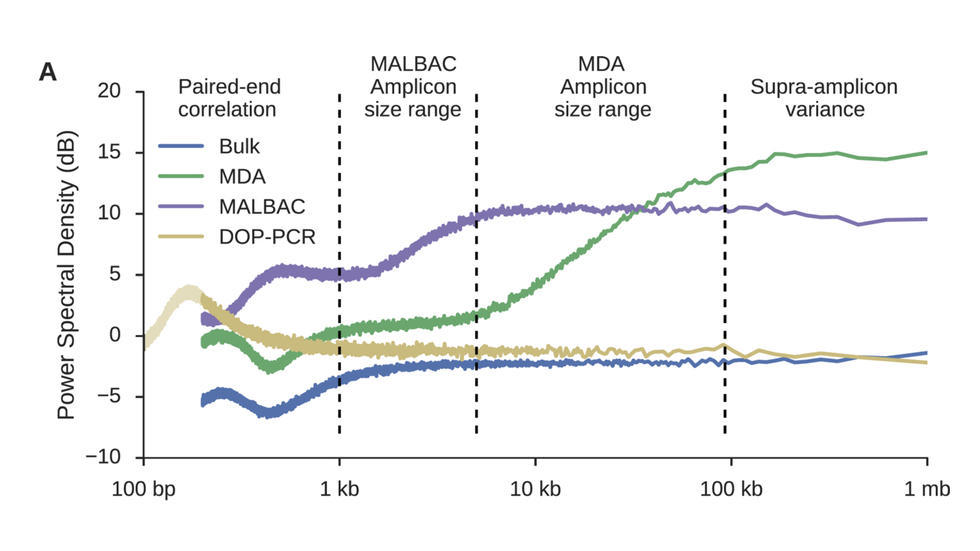
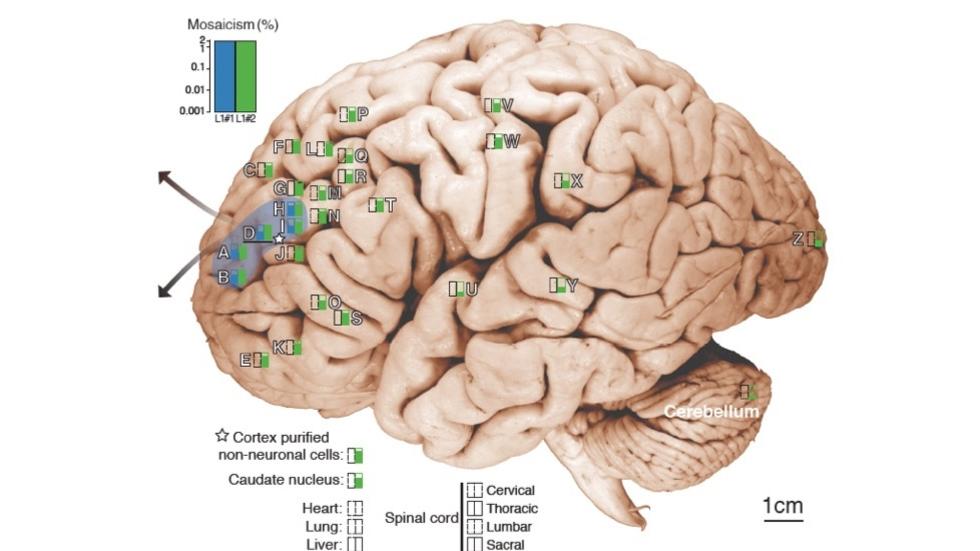
- Brain Somatic Mosaicism Network - The aim of this network is to characterize the spectrum of somatic variation in human brain samples and its role in psychiatric disorders. We are studying somatic mosaicism in autism with Chris Walsh (Boston Children's) and schizophrenia with Andy Chess (Mt. Sinai).
- Ludwig Center - This center brings together investigators from across Harvard to develop strategies to overcome the problem of therapy resistence in cancer.
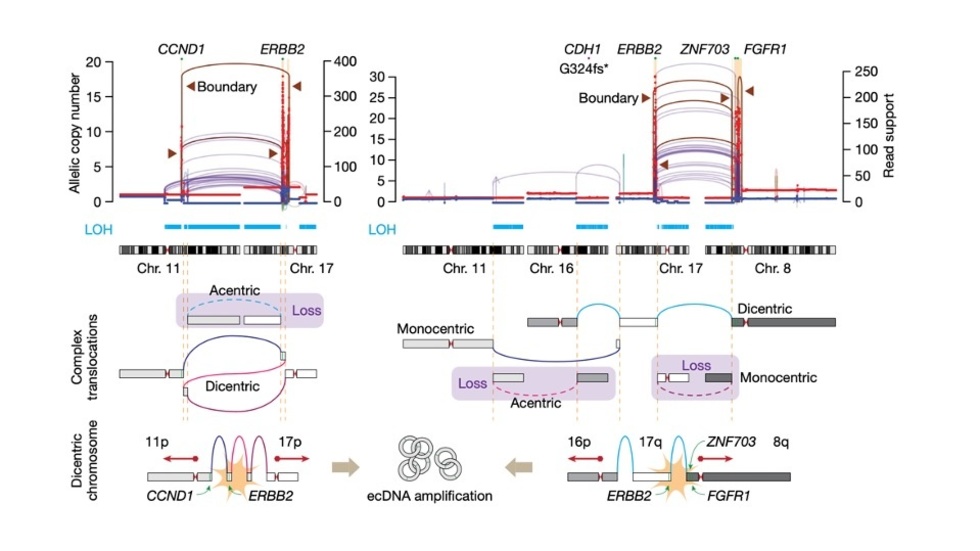
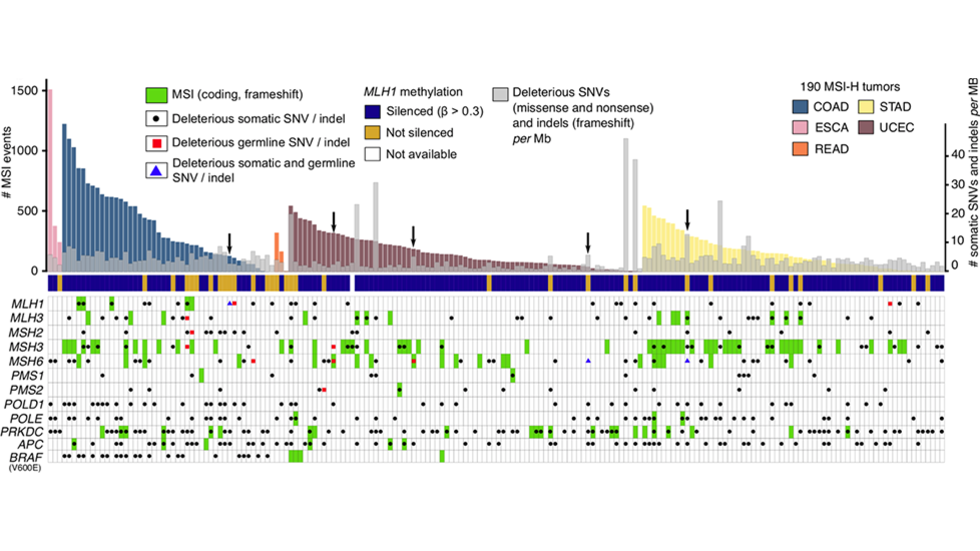
- International Cancer Genome Consortium - This is an effort by the international cancer genomics community to jointly analyze ~2,500 cancer genomes.